More Information
Submitted: February 20, 2024 | Approved: March 18, 2024 | Published: March 19, 2024
How to cite this article: Gazzah H, Hadrich Z, Tlili Y, Hafsi M, Hajri M, et al. Study on the Molecular Mechanism of Antioxidant Health Effect of Tuna Dark Meat Enzymatic Polypeptides. Arch Surg Clin Res. 2024; 8: 009-010.
DOI: 10.29328/journal.ascr.1001078
Copyright License: © Gazzah H, et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Keywords: Congenital cystic dilatation; Common bile duct; Total resection
Study on the Molecular Mechanism of Antioxidant Health Effect of Tuna Dark Meat Enzymatic Polypeptides
Zhang Diya3, Dong Xin2* and Guo Chunyu3
1Jiangsu Food and Drug Vocational Technical College, Experimental Training Management Center, China
2Lecturer, Master of Arts, School of Innovation and Entrepreneurship, Changzhou University, China
3Associate Researcher, Institute of Feed Research of Chinese Academy of Agricultural Sciences, China
*Address for Correspondence: Dong Xin, Lecturer, Master of Arts, School of Innovation and Entrepreneurship, Changzhou University, China, Email: [email protected]
In order to study the molecular mechanism of the antioxidant effect of enzymatically hydrolyzed tuna dark meat peptides, this article uses alkaline protease to enzymatically hydrolyze tuna dark meat, and at the same time performs peptide sequencing using matrix-assisted laser dissociation time-of-flight mass spectrometry (MALDI TOF/TOF). Discovery Studio (DS) performed molecular docking. Finally, the antioxidant effect was verified through DPPH clearance experiments. The results show that the dominant peptide sequences in the tuna dark meat hydrolyzed polypeptides are LAPGQ, GGGDPI, and PLRLP; through molecular simulation methods (Discover Studio, DS), the potential target of the above-mentioned enzymatic polypeptides was screened out to be Keap1, thus predicting antioxidant activity. It provides theoretical support for further research on enzymatic peptides. Through DPPH clearance experiments, it was found that both the enzymatic hydrolysate and LAPGQ, GGGDPI, and PLRLP have antioxidant activity, confirming their effects.
A large number of by-products are produced during tuna processing, and most of them are used in low-value applications and cause environmental pollution [1-4]. Effective enzymatic hydrolysis of tuna processing by-products through enzymatic hydrolysis technology to obtain bioactive peptides is an effective way to achieve high-value utilization of aquatic resources [5-7]. Han, et al. [8] studied the use of a combination of trypsin and alkaline protease to enzymatically hydrolyze dark tuna meat; Xin, et al. [9] used tuna minced meat protein as raw material and used trypsin and papain for enzymatic hydrolysis; it has been reported that Research reports have found that bioactive peptides derived from tuna have a variety of physiological activities. For example, Qian, et al. [10] found that the enzymatic hydrolyzate of bigeye tuna dark meat has antihypertensive activity; Hsu, et al. [11] found that the enzymatic hydrolyzate prepared from dark tuna meat has antioxidant effects. In order to elucidate the active mechanism of enzymatically hydrolyzed peptides, the mechanism of action of peptides can be analyzed through molecular docking methods [12-14]. Molecular docking predicts the interaction between ligands and receptors by calculating the interaction energy, binding sites, and other interaction information between molecules [15].
In this experiment, dark tuna meat was used as raw material and alkaline protease was used for enzymatic hydrolysis to obtain enzymatic peptides. The amino acid composition of the enzymatic peptides was sequenced and analyzed, and DS was further used to study the molecular mechanism. The health benefits of antioxidation were predicted, and the antioxidant function was verified through DPPH scavenging ability experiments, demonstrating its effectiveness.
Materials and reagents
Dark tuna meat was obtained from Food Co., Ltd.; alkaline protease (200,000 U/g) was purchased from Nanning Pangbo Bioengineering Co., Ltd.; a total amino acid (T-AA) detection kit was purchased from The Institute of Bioengineering was established in Nanjing; 1,1-diphenyl-2-picrylhydrazine (DPPH), N - [3- (2-furanyl) acryloyl] - L-phenylalanyl-glycidyl glycine (FAPGG), purchased from Sigma Corporation in the United States; Glutathione (GSH) was purchased from Beijing Solebao, and all other reagents are analytical grade.
5800 MALDI-TOF MS/MS instrument AB SCIEX Company of the United States; digital display constant temperature water bath HH-8 Changzhou Guohua Electric Co., Ltd. Discovery Studio software (Accelrys, USA).
Preparation of enzymatic peptides
Fresh dark meat is taken out of the refrigerator, washed with distilled water, and impurities are removed. Then, distilled water is added to a tissue homogenizer at a certain solid-liquid ratio and crushed to make a homogenization. The temperature and pH values are adjusted, and enzymes are added to a shaker for hydrolysis. The degree of hydrolysis is used as the evaluation index. The enzymatic hydrolysis temperature is 54.77 ℃, the amount of enzyme added is 2.86% and the enzymatic hydrolysis time is 4.49 hours. The enzyme activity is inactivated in a boiling water bath at 90 ℃ for 10 minutes and then centrifuged at 8000 r/min for 20 minutes. The supernatant is obtained, which is the enzymatic peptide solution.
Sequence analysis of enzymatic peptides
Perform sequence analysis of enzymatic peptides using MALDI TOF/TOF. Accurate molecular weight is determined by primary mass spectrometry, and amino acid sequence is determined by secondary mass spectrometry. Firstly, centrifuge the sample at high speed and remove the supernatant 40% μL. Dilute 20 times with water, take 1 μL sample, point on 5800 target plate, wait for drying, then add 1 more μL α- Cyano-4-hydroxycinnamic acid (CHCA) matrix, inject after drying. Collect and analyze the mass spectra of the samples, and compare them with the library using MASCOT software to determine the primary structure of the peptides.
Feature filtering based on discovery studio
Taking the dominant peptides in enzymatic hydrolysis as the research object, the peptide sequence was input into the Build and Edit Protein module of Discovery Studio (DS) to obtain the predicted 3D structure of the peptide, and the energy was optimized through the Full Minimization function in the Minimize Ligands module. Further, use the Pharma DB database and run the Ligand Profiler tool in the Pharmaphor module to screen target genes and predict peptide functions. Set the operation parameters to default values.
Molecular docking
Retrieve and download the three-dimensional structure of the screened target proteins in the PDB database, and use the Prepare Protein module of DS to process the proteins, dehydrate, hydrogenate, and optimize the protein structure. In Tools Explorer, Define Receiver defines the processed protein molecule as a receptor, defines the aforementioned peptides as ligands, and uses the CDOCKER tool for molecular docking.
Determination of DPPH radical scavenging activity
Synthesis of dominant peptides obtained through sequencing using solid-phase synthesis method as test samples. Mix 0.1 mL of the sample solution to be tested with 0.1 mL of prepared 0.1mmol/L DPPH methanol solution and shake well. After standing at room temperature and avoiding light for 30 minutes, measure its absorbance at 517 nm and calculate it as A1; Simultaneously measure the absorbance values of 0.1 mL of the sample solution to be tested and 0.1 mL of methanol, denoted as A2; And the absorbance value of 0.1 mL DPPH solution added to 0.1 mL distilled water is recorded as A0. Distilled water is used as a blank for control measurement. Glutathione (GSH) is used as a positive control. All experiments will be conducted in triplicate, with three parallel experiments in each group. Calculate the clearance rate of DPPH free radicals according to the formula:
DPPH clearance rate (%)=[1- (A1-A2)/10] x 100%
The concentration of the clearing agent in the test sample with a clearing activity of 50% is the semi-inhibitory concentration, denoted as IC50. The IC50 value is defined as the effective concentration of peptide molecules required to clear 50% of free radical activity.
Statistical analysis
Calculate IC50 through non-linear regression curve fitting using GraphPad Prism 5.0 (GraphPad Software Inc., San Diego, CA, USA).
Identification of peptide sequences
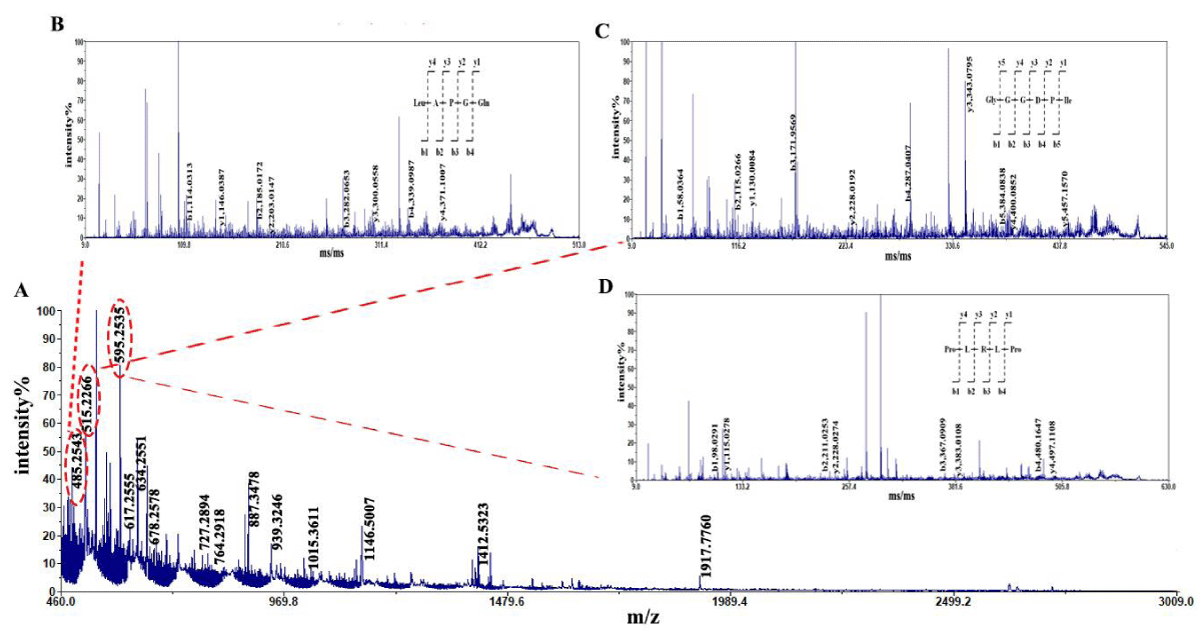
The amino acid sequence of the enzymatic hydrolysate was determined using MALDI-TOF/TOF. Figure 1 shows the results of primary mass spectrometry (MS) and secondary mass spectrometry (MS/MS) treatments. From Figure 1, it can be seen that the signal intensity of the mother ion m/z m/z 485.2453 is the highest in the peptide hydrolysis by alkaline protease, followed by the signal intensity of m/z m/z 515.2266 and m/z 595.2535, and the signal intensity of the other mother ions is relatively low. It can be considered that peptides with molecular weights of 485.2453 Da 515.2266 Da and 595.2535 Da are dominant in the enzymatic hydrolysate. Select them as parent ions for secondary mass spectrometry analysis, perform spectral library comparison on the obtained results, and obtain three dominant peptides with amino acid sequences of LAPGQ (Leu-Ala-Pro-Gly-Gln), GGGDPI (Gly-Gly-Gly-Asp-Pro-Ile) and PLRLP (Pro-Leu-Arg-Leu-Pro) (Table 1). Therefore, sequencing revealed that the molecular weight of peptides in the enzymatic hydrolysate was concentrated below 1kDa, and the dominant peptides in the alkaline protease hydrolysate were LAPGQ, GGGDPI and PLRLP.
Figure 1: Mass spectrogram of peptides from tuna dark muscle hydrolyzed by alkaline protease. Note: A: MS B: MS/MS of LAPGQ; C: MS/MS of GGGDPI; D: MS/MS of PLRLP.
| Table 1: The peptides identified in tuna dark muscle by enzyme. | |||||
| Note | [M + H] + | Area | % | Sequence | Kinds |
| 1 | 485.2453 | 80083.17 | 5.19 | LAPGQ | alkaline protease |
| 2 | 515.2266 | 74656.88 | 4.84 | GGGDPI | |
| 3 | 595.2535 | 98131.27 | 6.35 | PLRLP | |
Function prediction
Using the PharmaDB module for reverse targeting, the results showed that LAPGQ, GGGDPI, and PLRLP all interact with Kelch-like ECH-associated protein 1 (Keap1), as shown in Table 2.
| Table 2: Functional predictions of peptide sequences in tuna dark muscle. | ||||
| Note | Peptide sequence | login number | Pharmacophore abbreviation | Pharmacophore name |
| 1 | LAPGQ | P02829 | 1a4h | ATP-dependent molecular chaperone HSP82 |
| Q03518 | 1jj7 | Antigen peptide transporter 1 | ||
| P0C6X7 | 1uk4 | Replicase polyprotein 1ab | ||
| Q14145 | 5dad | Kelch-like ECH-associated protein 1(Keap1) | ||
| GGGDPI | P02829 | 1a4h | ATP-dependent molecular chaperone HSP82 | |
| P22637 | 1coy | Cholesterol oxidase | ||
| Q03518 | 1jj7 | Antigen peptide transporter 1 | ||
| P02766 | 1tyr | Transthyretin | ||
| 2 | P0C6X7 | 1uk4 | Replicase polyprotein 1ab | |
| P06241 | 2dq7 | Tyrosine-protein kinase Fyn | ||
| Q14145 | 5dad | Kelch-like ECH-associated protein 1(Keap1) | ||
| PLRLP | P60568 | 1m48 | Interleukin-2 | |
| P6WQG9 | 1m4d | Aminoglycoside 2'-N-acetyltransferase | ||
| P02766 | 1tyr | Transthyretin | ||
| O15151 | 3lbj | Protein Mdm4 | ||
| Q14145 | 5dad | Kelch-like ECH-associated protein 1(Keap1) | ||
Molecular docking of peptide molecules with Keap1
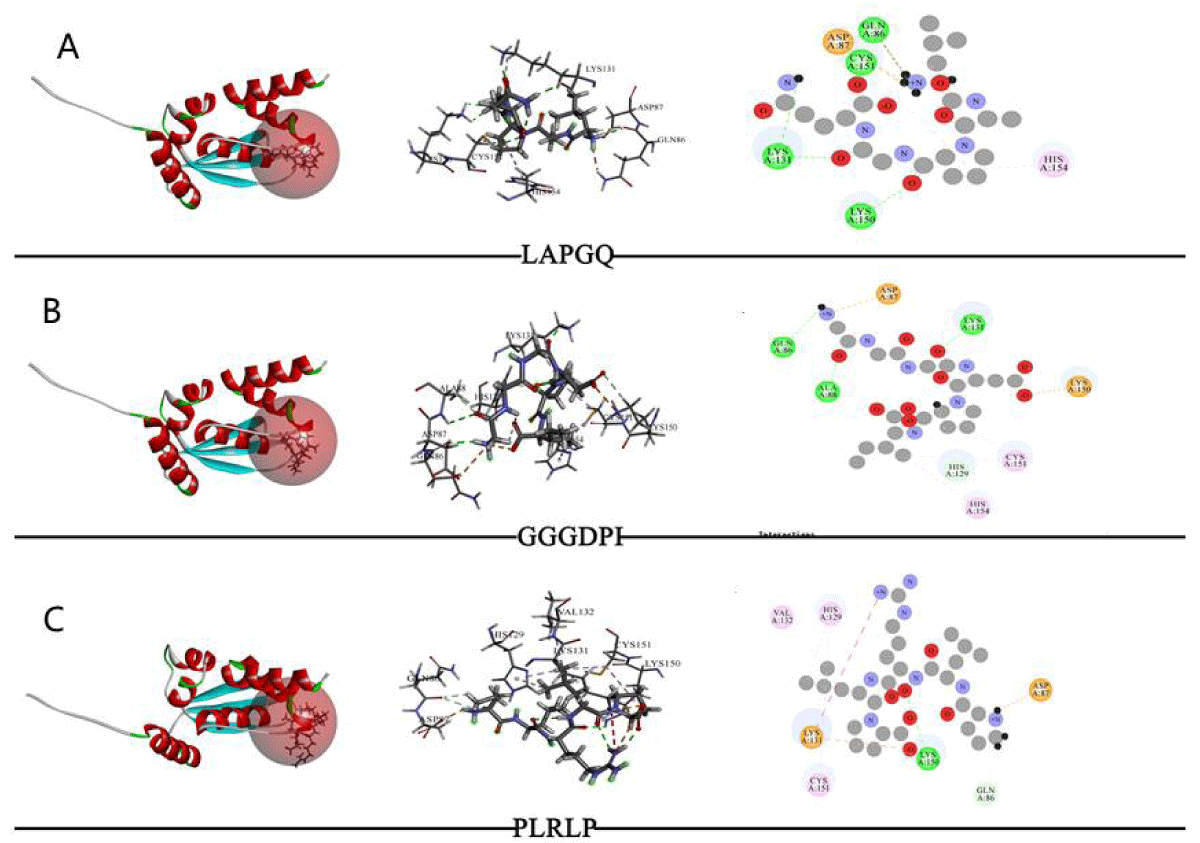
The molecular docking results of Keap1 with various peptides (LAPGQ, GGGDPI, PLRLP) are shown in Table 3 and Figure 2. The results of CDocker molecular docking mainly determine the degree of binding based on information such as the energy of the receptor-ligand (- CDOCKER Energy, - Ec), electrostatic energy (Eele), and Van der Waals Energy (Evdw). The value of - CDOKER-Energy also indicates the strength of the affinity between peptides and receptor proteins. According to Table 3, GGGDPI (86.4534 kcal/mol) showed higher affinity than LAPGQ (68.8912 kcal/mol) and PLRLP (38.4365 kcal/mol). From Figure 2, it can be seen that LAPGQ interacts with Keap1 with a total of 6 amino acid residues, forming 4 HBs with Gln86, Lys131, Lys150, and Cys151; There are a total of 8 amino acid residues in the interaction between GGGDPI and Keap1, forming 3 HBs with Gln86, Ala88, and Lys131; There are a total of 7 amino acid residues involved in the interaction between PLRLP and Keap1, forming HB with Lys150.
| Table 3: The results of molecule docking between peptides and Keap1. | |||||
| Sequence | -Ec | Eele | Evdw | Dooker | Interacting amino acid residues |
| LAPGQ | 68.8912 | -191.164 | -12.4822 | 1 | 6 |
| GGGDPI | 86.4534 | -302.939 | -9.60284 | 1 | 8 |
| PLRLP | 38.4365 | -206.416 | -15.6822 | 1 | 7 |
Figure 2: The results of molecule docking between peptides and Keap1. Note: From left to right, the docking status of active sites, amino acid residue connections, and molecular docking two-dimensional diagram.
DPPH radical scavenging activity of peptides
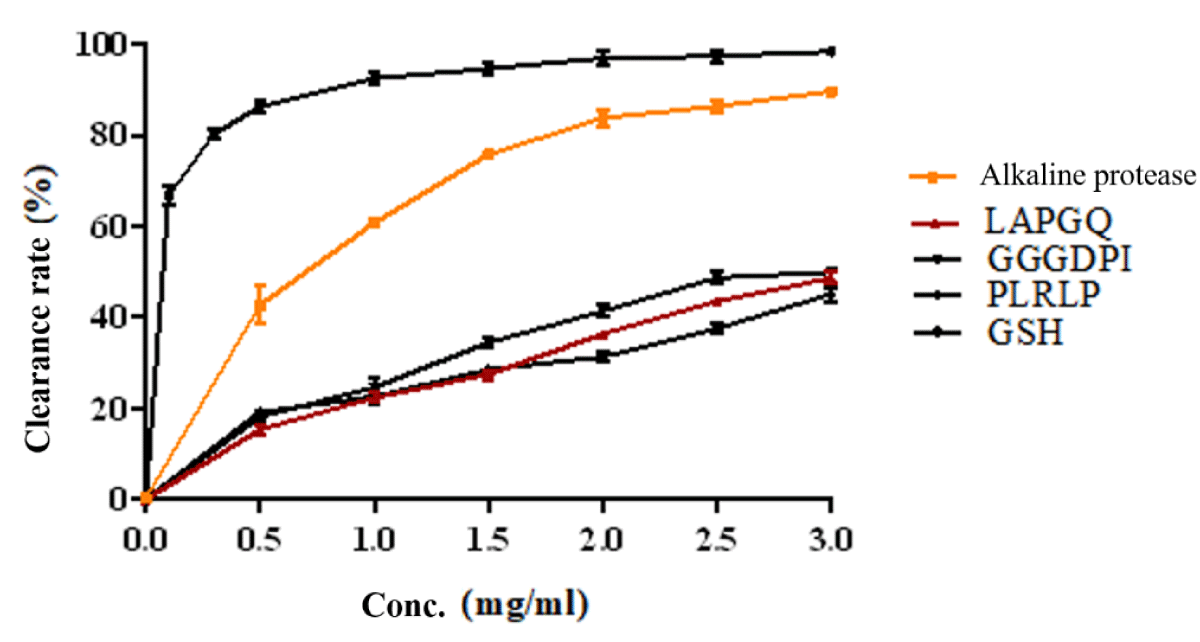
The scavenging rate of DPPH free radicals and the antioxidant activity of enzymatic hydrolytic peptides from dark tuna meat are shown in Figure 3. The results showed that both the enzymatic solution and synthetic peptides under enzymatic conditions exhibited antioxidant activity. The IC 50 values of alkaline protease, LAPGQ, GGGDPI, and PLRLP were 0.6588 mg/ml, 3.358 mg/ml, 2.900 mg/ml, and 4.861 mg/ml, respectively. This indicates that the enzymatic hydrolysate has better antioxidant activity.
Figure 3: DPPH scavenging activities of synthesized peptides.
This article focuses on the functional prediction of dominant peptides after sequencing the enzymatic hydrolysis peptides of dark tuna meat. Keap1 protein is a multi-domain repressor protein mainly present in the cytoplasm, which can inhibit Nrf2 activity. Usually, Nrf2 is combined with its inhibitor Keap1 and exists in an inactive state in the cytoplasm. However, when cells are stimulated by reactive oxygen species, Nrf2 decouples from Keap1, and activated Nrf2 is transported into the nucleus, activating target gene expression and regulating the transcriptional activity of phase II metabolic enzymes, antioxidant enzymes, or drug transporters, thereby exerting antioxidant damage [16,17]. Huerta, et al. [18] found that TX6 can bind to Keap1 protein, thereby inhibiting Keap1 activity and activating the Nrf2/ARE signaling pathway. The molecular docking results of this study showed that in the alkaline protease-treated group, three dominant peptides interacted with Keap1 through common amino acids Lys131, Asp87, and Lys150, which played an important role in the binding process. Thus, it can be predicted that the binding of enzymatic peptides to Keap1 protein will inhibit Keap1 protein activity, affect the Keap1-Nrf2/ARE signaling pathway, and play an antioxidant role.
Finally, the DPPH radical scavenging activity of the enzymatic hydrolysate and synthesized peptide molecules was validated through in vitro experiments. The results showed that both enzymatic hydrolysate and synthetic peptides had varying degrees of antioxidant activity, confirming the correctness of molecular docking and computer simulation.
- Herpandi NH, Rosma A, Nadiah WAW. The tuna fishing industry: a new outlook on fish protein hydrolysates. Comprehensive Reviews in Food Science & Food Safety. 2011; 10(4): 195-207.
- Sohn JH, Ohshima T. Control of lipid oxidation and meat color deterioration in skipjack tuna muscle during ice storage. Fisheries Science. 2010; 76(4): 703-710.
- Su Y. Analysis of Nutrient Composition and Study on Anti fatigue Activity of Muscle Protease Hydrolysates of Three Tuna Species from the South China Sea. Guangdong: Guangdong Ocean University, 2015.
- Su Y, Zhang C H, Cao WH. Analysis and Evaluation of Nutritional Composition of Three Kinds of Tuna Common Meat and Dark Meat Produced in the South China Sea [J] Journal of Guangdong Ocean University. 2015; 35(3):87-93.
- Je JY, Qian ZJ, Byun HG. Purification and characterization of an antioxidant peptide obtained from tuna backbone protein by enzymatic hydrolysis. Process Biochemistry. 2007; 42(5): 840-846.
- Je JY, Lee KH, Mi HL. Antioxidant and antihypertensive protein hydrolysates produced from tuna liver by enzymatic hydrolysis. Food Research International. 2009; 42(9): 1266-1272.
- Liu JH, Wang B, Kuai P. Enzymatic hydrolysis process of dark tuna meat and nutritional value evaluation of its hydrolysates. Food Science. 2014; 35(20): 1-5.
- Han J, Tang S, Li Y. In silico analysis and in vivo tests of the tuna dark muscle hydrolysate anti-oxidation effect. Rsc Advances. 2018; 8(25): 14109-14119.
- Xin J M, Wang P, Li S H. A Study on the Technology of Compound Enzymatic Hydrolysis of Broken Tuna Meat. The Food Industry. 2011(9); 25-28.
- Qian ZJ, Je JY, Kim SK. Antihypertensive effect of angiotensin i converting enzyme-inhibitory peptide from hydrolysates of Bigeye tuna dark muscle, Thunnus obesus. J Agric Food Chem. 2007 Oct 17;55(21):8398-403. doi: 10.1021/jf0710635. Epub 2007 Sep 26. PMID: 17894458.
- Hsu KC. Purification of antioxidative peptides prepared from enzymatic hydrolysates of tuna dark muscle by-product. Food Chemistry. 2010; 122(1): 42-48.
- Park K, Cho AE. Using reverse docking to identify potential targets for ginsenosides. Journal of Ginseng Research.
- Grienke U, Kaserer T, Pfluger F, Mair CE, Langer T, Schuster D, Rollinger JM. Accessing biological actions of Ganoderma secondary metabolites by in silico profiling. Phytochemistry. 2015 Jun;114:114-24. doi: 10.1016/j.phytochem.2014.10.010. Epub 2014 Nov 6. PMID: 25457486; PMCID: PMC4948669.
- Xie CL, Chong SY, Cao GP, In silico investigation of action mechanism of four novel angiotensin-I converting enzyme inhibitory peptides modified with Trp. Journal of Functional Foods. 2015; 17: 632-639.
- Tu M, Wang C, Chen C, Zhang R, Liu H, Lu W, Jiang L, Du M. Identification of a novel ACE-inhibitory peptide from casein and evaluation of the inhibitory mechanisms. Food Chem. 2018 Aug 1;256:98-104. doi: 10.1016/j.foodchem.2018.02.107. Epub 2018 Feb 21. PMID: 29606478.
- Gao B, Doan A, Hybertson BM. The clinical potential of influencing Nrf2 signaling in degenerative and immunological disorders. Clin Pharmacol. 2014 Feb 3;6:19-34. doi: 10.2147/CPAA.S35078. PMID: 24520207; PMCID: PMC3917919.
- Hu LF, Wang Y, Ren RJ. The antioxidant stress effect and regulatory mechanism of Keap1-Nrf2/ARE signaling pathway [J] International Journal of Pharmaceutical Research. 2016; 43(1): 146-152.
- Huerta C, Jiang X, Trevino I, Bender CF, Ferguson DA, Probst B, Swinger KK, Stoll VS, Thomas PJ, Dulubova I, Visnick M, Wigley WC. Characterization of novel small-molecule NRF2 activators: Structural and biochemical validation of stereospecific KEAP1 binding. Biochim Biophys Acta. 2016 Nov;1860(11 Pt A):2537-2552. doi: 10.1016/j.bbagen.2016.07.026. Epub 2016 Jul 27. PMID: 27474998.